Some of the more interesting developments include the consideration of the phenomenon of Spiral Phyllotaxis, aging models for the decomposition of plants over time, ecosystem models for simulating whole fields of wildflowers, and a modular approach to a symbiotic system where environmental parameters affect plant development and plant development effects the environment. A quick survey of work done follows.
In 1992, Deborah Fowler investigated the use of collisions while implementing the process of Spiral Phyllotaxis as a guideline to creating virtual plant models [8]. Phyllotaxis is a well-documented phenomenon in plants that determines where primordia (seeds, petals, nuggets etc.) are located on a recepticle (sunflower, coneflower, corn ear) using a fixed divergence angle of 137.5 degrees. Primordia are packed following the rules of phyllotaxis until they collide with each other. Phyllotaxis has been incorporated into all plant development simulation systems as a result of Fowler and others’ work.
In 1996, Radomir Mech devised a system design where virtual plants could be modularly developed through interaction with other environmental modules [9]. Using such a system, virtual plants don’t grow in a vacuum, but instead have their parameters affected by dynamic environmental parameters. For example, if the environment experiences a drought, plant growth is stunted and body structures wilt. The communication goes in both directions. If a species of plant becomes successful in growing and respiring, additional oxygen is passed back to the environment.
In 1998, Oliver Deussen led a team that focused on realistic modeling of plant ecosystems [10]. Starting with a terrain specification and environmental characteristics such as soil humidity, the researchers’ virtual plant engine distributed plant species across the terrain. With the engine running on a supercomputer array, a computer interface afforded a user the ability to zoom in and zoom out while investigating the component wildflowers that had grown to fill the terrain.
The Vehicles of Evolution
Once procedural plant models are driven by parameters to a virtual plant growth engine, nature can provide additional inspiration in evolving new plants from existing parameter lists. The process mimics nature by repackaging parameter lists into psuedo genomes that then simulate physical plant genomes. Once the parameters are in a genome format, the forces of evolution can be written algorithmically in the virtual plant application engine to provide a variety of plant offspring.
Chris Colby of Boston University classifies observed phenomena of genome evolution into five processes: mutation, recombination, gene flow, genetic drift, and natural selection [11]. All five processes can be simulated in lines of code. A quick mention of each is warranted here.
Mutation occurs when some external process (such as a cosmic ray) interferes with the normal functioning of DNA within live cells and forces a change in its chemical makeup. The changed structure then can change the mutated structure’s function. Mutation usually does more harm than good to the affected entity but in nature, on average, only about 1 x 10-11 mutations occur per base pair of DNA per generation.
Recombination occurs most spectacularly through meiosis during sexual reproduction. During meiosis, the mother provides half the offspring’s genetic material and the father provides the other half. So, the next generation’s DNA varies quite dramatically from that DNA of the mother or father’s individual.
Gene Flow occurs when some process (such as a mosquito drawing blood) transfers genetic material from one species to another and that material makes its way into the DNA of the target species. The process has been documented between species of fruit flies with a parasitic mite as carrier.
Genetic Drift occurs as a purely statistical process. As a gene pool reproduces and survives in an environment where death is caused by events outside its control, chance determines which genes continue to reproduce and which genes die out.
Natural Selection refers to the process where a gene pool's makeup changes based on reproducing and surviving in an environment where the individual does have some control over their survival. Those individuals with genes that are better suited to the environment tend to reproduce more often than those less suited. Volumes have been written about the sub processes of natural selection: ability to feed, ability to avoid predators, sexual selection, etc. as Charles Darwin intrigued the world with his study of life’s struggle in the Galapagos Islands of South America.
My Contribution
To further along research in the area of modeling virtual plants, I chose to make three contributions: to enhance a popular L-Systems engine to take virtual plant genomes as input, to add a vase genome to the plant model to suggest use in artistic expression, and to explore gene pool variety using lessons from evolutionary process. The remainder of this paper explains work done and provides results and discussion.
Enhancing the LParser Engine to Take Virtual Plant Genomes As Input

|
Figure 1 - Rendered VRML97 output of Flowers1.ls |
I dissected the Flowers1.ls description file to determine which statements (also called rules) in the file influenced which part of the final plant model form. My conclusions were as follows:
Description File Rule . . . General Conclusion
|
q q=&(30)p^(30)p^(10)p p=[t(-0.2)!Ft(-0.2)!Ft(-0.2)!Fo] o=>y>(60)y>(60)y>(60)y>(60)y>(60)>y a=[&(10)s]a y=[a&(125)w] w=Z[d][h]'^(30)cw d=[+d{.].x.} h=[^(5)-h{.].x.} x=^(5)ggx s=[c(5)^(60)!'Ft(0.6)ZZt(0.6)?(10)ZZ] F='(1.3)F'(.77) |
The Axiom (all descriptions begin with the Axiom) q sets the number of stalks and their placement p creates the stalk (with t providing a gravity force) o sets the number of petals around the flower a sets the overall shape of the petal y is a component of the petal shape w is a component of the petal shape d is the right hand side of the petal h is the left hand side of the petal x pushes the vertex formation of the petal forward s creates the stamen F directs the overall size of each forward instruction |
Because L-Systems incorporate recursion, the processes by which petals and leaves are generated are similar with the difference being that leaves are created in earlier iterations and are colored green, while petals are created in later iterations and are brightly colored at the red and violet ends of the color spectrum (not that virtual plants have to conform to the stereotypes of physical plants). Thinking that through, I realized that I could add an additional set of rules to the Flowers1.ls description file that would generate the plant leaves independently of the flower petals.
I then generalized the description file to substitute variables where fixed coefficients had been used by C.J. and added interesting features from other description files in order to provide a wider possibility of form from the genome input engine. I put component plant parts into for loops to be able to vary the number of parts that would be created. For example, to calculate the number of stalks in the model, I wrote the following snippet of code
for(counter=1;counter<=numstalks;counter++) {
strcat(rule[0], "^(20)S");
}
where the first rule would concatenate one stalk (represented by S for each additional stalk up to the number of stalks stored in the model genome. I then derived the positioning of the next stalk from the previous (represented by ^ from another value stored in the genome. I continued adding coding branches and loops, replacing coefficients with variables, and adding new features from other L-System description files until I had produced a rich enough system to let each component of my plant genome affect a wide-range of possible plants.
An example plant genome is stored as a single line text string like:
YY|NN|3|0.3|0.3|0.0|0.5|YY|NN|YN|NY|2|4|YY|NN|YN|NY|3|0.2|0.6|0.15|10|1|YY|NN|YN|NY|4|0.6|0.2|0.1
where each pipe symbol separates an allele pair or simulated gene from its neighbors. Form that is driven by dominant or recessive alleles is stored as a series of NN, NY, YN, or YY pairs where Y is the dominant trait and N the recessive. Form that supports color is stored in three floats contributing red, green, and blue components of light in that order. All other values are single floats or integers that substitute into rule variables in the virtual plant generation process.
Figure 2 lists the mapping of the stored genome by responsibility during virtual model development. The genome focuses on three of many potential plant body parts: The stalk; the leaves, and the petals.

|
Figure 2 - My Virtual Plant Genome |
Adding a Vase Genome
In order to differentiate my work from those who thoroughly understand botany, I decided to create a vase genome as well to suggest application in creating objects of artistic expression. Virtual plants need not be realistic to be interesting or entertaining. My vase genome is stored as a simple single-lined text string like:
750^25^50^75^100^-90^-80^-70^-60^-50^-40^-30^-20^-5^5^5^5^5^5^5^5^Male^0.8^0.8^0.2^1.0^0.8^0.2^YN
where each value is substituted into a vase generation process I added to the LParser engine. Female vases are completely circular in any top view cross-section while male vase top view cross-sections have corners. Much of the vase genome simply stores the radius of the vase at different heights in the model. The two sets of RGB color values at the end are identical if the vase carries two recessive alleles for stripes (and then the second triplet is ignored). The complete vase genome is shown in Figure 3.

|
Figure 3 - The Vase Genome |
Exploring Gene Pool Variety in Virtual Plants
Having succeeded at generating 3-D plant models from plant genomes, I decided to put in place a process to generate new plant genomes from existing plants. I added three new processes to simulate gene pool processes described earlier.
To implement mutation, I represented each numeric value in the genome as a base two binary number and flipped random bits. An interesting example was one where an orange petal genome and red petal genome produced next-generation petals that were brown. I implemented an averaging process (with mild noise) to create offspring petal color and looking at the results came to the conclusion that the most-significant bit of the child petal’s red color component had been flipped from 1 to 0 (thus reducing the amount of red dramatically).
To implement recombination, I created a merging process where two plant genomes could come together to create a new plant genome. Those traits driven by dominant and recessive alleles simulated meiosis by each randomly providing one of the two base alleles to the next-generation plant. Non-allele specified traits were merged using an averaging process with noise. I adjusted the noise threshold such that most of the time the offspring trait value would fall between the range of both parents. Yet, values outside of the range (especially for narrow ranges) were statistically possible at two standard deviations from the norm.
To implement genetic drift, I ran multiple generations and watched for drift. When values drifted toward too narrow a value (symptomatic of poorly designed code) consistently, I reworked the engine to provide a wider range of potential drift.
In considering Stephen Rooke's process for genetically evolving art paintings, I agreed human like and dislike were representative forms of environmental pressure simulating natural selection [13]. Although I haven't personally applied my aesthetic as a genetic pruning mechanism, I watch the process in action when I installed my engine at a booth during April 21-May 6, 2001 at the Boston Cyberarts Festival [14] in Boston, MA. More than 700 visitors to the booth were able to mate two virtual plants and decide if the offspring should survive to mate themselves.
Visitors experienced the plants by using the University of Washington HIT Lab's Magic Book technology. The Magic Book allows for a mixed reality presentation so visitors will be able to see the 3-D virtual plants overlaid on video processed from their real world perspective [15]. Figure 4 shows a participant placing two plants together to mate them at the Boston Architecture Center exhibition on April 26, 2001.

|
Figure 4 - Mating Two Plants at the Boston CyberArts Festival |
Examples
Figure 5 shows the visual effects of recombination from alleles that define leaves on stem or not. The first two columns show the visual effects of the allele contribution from both parent plants. The third column shows the visual effect in the child. Since having leaves on the stem is a dominant trait in the system, a plant must possess two recessive alleles in order to display the recessive trait (lack of leaves on the stem).
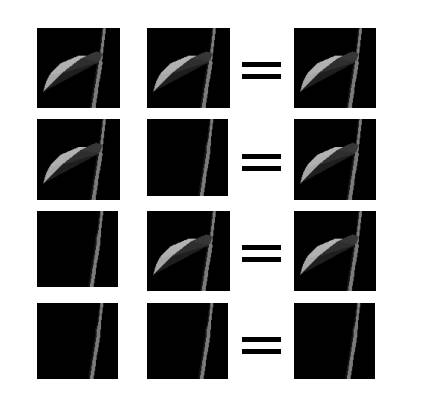
|
Figure 5 - Effect of Recombination of Alleles on Stem Leaves |
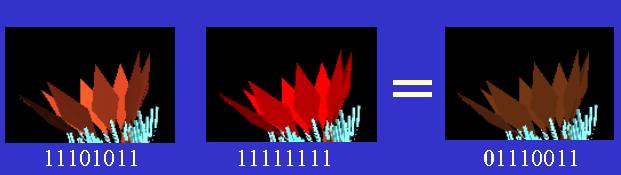
|
Figure 6 - Example of Mutation in the Red Color Component |

|
Figure 7 - A 3-D Model Created by the Engine |
Conclusions and Future Work
I found playing with parametric model generation engines intoxicating and innately creative when the goal was not much more than exploration. Many of the decisions I made and implemented in code are arbitrary in their purpose. The process felt more like artistic expression than scientific discovery and yet the process was inspired by readings in the sciences. Though there are 1020 possible models generated by my simple genomes (before applying noise algorithms), I still have only touched an infinitesimal percentage of all possible plants generated from L-System rules. Playing with L-Systems has taught me to more deeply appreciate nature and the variety life process produces without any necessary conscious guidance.
Speculation that evolutionary process can evolve new processes in the digital realm appears to be a hot topic based on the number of books one can find related to the subject. Investigators such as Tom Ray of the University of Oklahoma continue to opine that great things will come of digital evolution seeded properly and left to its own device [16]. Such popular science discussion often overshadows the significance of what we already know how to do. Generating a wide variety of complex 3-D models to enhance our virtual environments in a real-time system seems to be a worthwhile goal on its own accord.
Through my explorations for this paper, I have gained a deeper understanding of how digitally produced movie sets, such as the one used recently in The Grinch Who Stole Christmas, will continue to evolve to become more interesting and more realistic at the same time. If variety truly is the spice of life, these parametric modeling engines are worth their weight in gold. As long as bandwidth continues to be a limiting bottleneck in real-time, shared virtual environments, research in this field should be considered important and invaluable. Other processes of nature in the chemical and physical sciences should be reaped as inspirational models for engine development.
Certainly both the plant and vase genomes could continue to be expanded to enact an even finer level of control during virtual plant generation. But, perhaps, improvement of the engine itself should come first. Currently, the engine produces plant body parts that have no mass and thus can intersect in unfortunate places. Often, the output in that case appears immediately ugly to the eye. Sometimes, the effect is desirable, especially when the intersecting polygons are near the same plane or are of the same color. Collision detection techniques that are well understood algorithmically today could be incorporated to preclude those situations where the virtual plant immediately appears unattractive.
Of final interest are the comments made by participants who mated plants at the Boston CyberArts' Festival. Suggestions participants were especially keen on seeing realized included:
- Applying the genome from a grid of the Magic Book patterns whereby each zone of the pattern could directly represent the state of the genome (and where participants could draw patterns at the exhibit).
- Networking the application so mulitiple participants could adjust the patterns across the Web to see what plants would result.
- Running the engine on a higher performance platform so the plants could appear more realistic.
- Applying the engine to avatar generation techniques (using the general public as bases for genomes).
References:
[1] Schwartz, Paul et al, Virtual Playground:
Architectures for a Shared Virtual World, http://www.hitl.washington.edu/publications/r-98-12/
(Accessed 12 February, 2001)
[2]
ActiveWorlds, Welcome To The 3D Internet, http://www.activeworlds.com
(Accessed 24 February, 2001)
[3]
Przemyslaw Prusinkiewicz et al, Visual models of plant development. In G.
Rozenberg and A. Salomaa, editors, Handbook of formal languages.
Springer-Verlag, 1996. To appear.
[4]
Karl Sims, Evolving Virtual Creatures, Proceedings of SIGGRAPH ’94
(Orlando, Florida, July 24-29, 1994) pp 15-22. [5] MendelWeb, Mendel’s Paper, http://www-hpcc.astro.washington.edu/mirrors/MandelWeb/
(Accessed 26 February, 2001)
[6] Aristid Lindenmayer, Mathematical Models for Cellular
Interaction in Development, Parts I and II. J. Theoretical Biology,
18: (1968), pp. 280-315.
[7] The University of Calgary Web Server, Biological Modeling
and Visualization, http://www.cpsc.ucalgary.ca/Research/bmv/index.html
(Accessed 24 February, 2001)
[8] Deborah R.
Fowler, Przemyslaw Prusinkiewicz, and Johannes Battjes A Collision-based Model of Spiral Phyllotaxis.
Proceedings of SIGGRAPH '92 (Chicago, Illinois, July 26-31, 1992), In Computer
Graphics, 26, 2, (July 1992), ACM SIGGRAPH, New York, pp. 361-368.
[9] Radomir Mech and Przemyslaw Prusinkiewicz Visual Models of
Plants Interacting with Their Environment. Proceedings of SIGGRAPH '96 (New
Orleans, Louisiana, August 4-9, 1996).
[10] Oliver Deussen et al. Realistic Modeling and Rendering of
Plant Ecosystems. Proceedings of SIGGRAPH '98 (Orlando, Florida, July
19-24, 1998).
[11] Chris Colby, Introduction to Evolutionary Biology, http://www.talkorigins.org/faqs/faq-intro-to-biology.html
(Accessed 25 February, 2001)
[12] Laurens Lapre, LParser, http://www.xs4all.nl/~ljlapre/lparser.htm
(Accessed 20 January, 2001)
[13] Steven Rooke, The Genetic-Evolutionary Art Process
Employed by Steven Rooke, http://www.azstarnet.com/~srooke/process.html
(Accessed 14 February, 2001)
[16] Boston Cyberarts Festival, Boston Cyberarts Festival, http://www.bostoncyberarts.org/ (Accessed
1 March 2001)
[15] Mark Billinghurst, The Magic Book, http://www.hitl.washington.edu/magicbook/
(Accessed 1 March, 2001)
[16] Tom Ray, Beyond The Turing Test, Lecture at The
Digital Biota 3 OWorlds Conference (San José, CA, November 6-7, 1999)